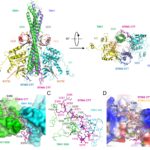
The detection of pathogen-derived nucleic acids is a central strategy by which the host senses infection and initiates protective immune responses. Cyclic GMP-AMP synthase (cGAS) is a dsDNA sensor. It catalyzes the synthesis of cyclic GMP-AMP (cGAMP), which stimulates the induction of type I interferons through the STING–TBK1–IRF-3 signaling axis. However, the precise mechanisms that govern activation of STING by cGAMP and subsequent activation of TBK1 remain unclear. A paper published by Dr. Pingwei Li’s… Read More →
Finding cancer-promoting proteins in bacteria
Researchers in the Hu lab created the online data browser for the E. coli-to-Cancer Gene-function Atlas (ECGA), which was described in the Jan 10, 2019 issue of Cell. “Our cells make protein carcinogens,” said co-corresponding author Dr. Susan M. Rosenberg, Ben F. Love Chair in Cancer Research and professor of molecular and human genetics, of molecular virology… Read More →
Charlie Farber
Charlie Farber is a second year graduate student in the laboratory of Dr. Dmitry Kurouski. He is investigating the mechanisms of host/pathogen interactions using vibrational spectroscopy methods including Raman and IR. He is also interested in exploring the viability of Raman as a method for disease detection. Currently in his second year of the program, Charlie has published a… Read More →
Dr. Allison Cockrell Zugell
Dr. Allison Cockrell Zugell is a Scientist at Janssen, the pharmaceutical company of Johnson & Johnson. Her research group develops HPLC and capillary based methods for late-phase biotherapeutics. Allison received her PhD in 2013 in the lab of Dr. Paul Lindahl. From 2013-2015, she worked at the Naval Research Lab in Washington, DC as a… Read More →
Dr. Mikaela Stewart
Dr. Mikaela Stewart received her PhD in 2013 in the lab of Dr. Tatyana Igumenova. She is an Assistant Professor at Texas Christian University. Mikaela teaches biochemistry and manages a lab with undergraduate and graduate students. Her research focuses on characterizing protein-protein interactions within BRCA1 intrinsically disordered region that are important for tumor-supression. Mikaela was… Read More →
Dr. Aaron Pawlyk
Dr. Aaron Pawlyk received his PhD in 2001 in the lab of Dr. Donald Pettigrew. He is a Program Director at the National Institute of Diabetes and Digestive and Kidney Diseases at the National Institutes of Heatlh. His job responsibilities include oversight of grants going to universities, scientific management of large NIH-funded consortia, and providing… Read More →
Dr. Julian Avila-Pacheco
Dr. Julian Avila-Pacheco received his PhD in 2012 in the lab of Dr. Tim Devarenne. He is a research scientist in the Metabolomics Platform of the Broad Institute of MIT and Harvard. Julian works on developing methods for the analysis and interpretation of metabolomics data generated using liquid chromatography tandem mass spectrometry (LC-MS). He works… Read More →
Yuan Zhi
Yuan Zhi is a 5th year student conducting research in the lab of Dr. Frank Raushel. He is interested in understanding how a carbohydrate transportation and metabolism related pathway play a role in Salmonella colonization in the gut of cattle. He has received three InnoCentive Winning Solvers Awards totaling $11,500. In August Yuan will be attending… Read More →
Natalie Garza
Natalie Garza is a third-year graduate student in the laboratory of Dr. Vishal M. Gohil. The goal of her project is to identify and characterize proteins involved in regulating intracellular copper transport to the mitochondria. The identification of these pathways will allow for better treatment options for metabolic disorders associated with copper deficiency. Natalie has… Read More →
Borja Barbero Barcenilla
Borja Barbero Barcenilla is a 4th year PhD student in the lab of Dr. Dorothy Shippen. He is investigating the moonlighting functions of telomere associated protein on Arabidopsis thaliana. He placed second in the oral presentation at the 20th annual Student Research Week (SRW). Borja is a former NCAA Division 1 athlete and holds records… Read More →